-
Interaction
| AccNo. |
22635 |
Score |
0.86 |
| Name |
DPRSA_LNCaP |
Environment |
in vitro / culture |
| Kd |
1.0 |
Organism |
Homo sapiens (human) |
|
Peptide
| AccNo. |
22519 |
| Name |
DPRSA |
| Organism |
N/A |
| Constraint |
none |
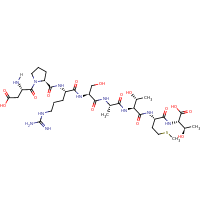
| Sequence |
DPRSATMT
|
| Origin |
phage display |
| Form |
phage |
| Internalized |
no |
| Unnatural |
no |
| Imaging |
no |
| Is Motif |
no |
|
Interactor
| AccNo. |
22361 |
| Name |
LNCaP |
| Description |
LNCaP prostate carcinoma cells |
| Organism |
Homo sapiens (human) |
| Type |
cell |
|
Experiment
| AccNo. |
22580 |
Classification incorrect?
Click the corresponding button to vote. Your vote
is used to improve the automatic classification.

|
| CA |
CVD |
DM |
APO |
ANG |
MI |
BD |
|
| 0.68 |
0.60 |
0.56 |
0.57 |
0.55 |
0.61 |
0.59 |
Vote |
 |
 |
 |
 |
 |
 |
 |
Yes |
 |
 |
 |
 |
 |
 |
 |
No |
|
| Name |
PD_8 |
| Detection |
filamentous phage display, MI:0048 |
| Source |
PDF |
Text Id |
8 |
| Journal |
Prostate. 2001 Jun 1;47(4):239-51. |
| Title |
Phage display selection of peptides that affect prostate carcinoma cells attachment and invasion. |
| Authors |
Romanov VI, Durand DB, Petrenko VA |
| Text |
BACKGROUND: Prostate cancer-specific proteins must be identified to serve as diagnostic and prognostic markers. Cell surface proteins are especially important, because they have potential utility as diagnostic markers and therapeutic targets. Identification of ligands for these proteins will allow use of these ligands as diagnostic and therapeutic tools and permit the investigation of receptor function. We performed a search for peptide ligands to prostate cancer cell-specific receptors. METHODS: Peptide phage display library was used to isolate specific ligands to LNCaP prostate carcinoma cells receptors. Selected phage and cognate peptides were investigated for their cancer-related functions, such as the ability to interfere with cell adhesion, spreading, motility, and invasion. RESULTS: Phage designated pg35, blocked spreading of LNCaP cells and their derivatives C4-2 and C4-2b. Cognate peptide did not inhibit spreading, but incubation of C4-2 and C4-2b cells with cognate peptide increased their affinity for endothelial cells and invasiveness. In addition, the peptide activates matrix metalloproteinase (MMP)-2 and 9 in C4-2 and C4-2b cells. CONCLUSIONS: These results indicate that identified ligands may play a role in tumorigenicity and metastatic transformation of prostate cancer. To our knowledge, this is the first identification of a functional cancer-specific peptide ligand using the phage display approach. |
| Mesh Terms |
Amino Acid Sequence; Animals; Bacteriophages/metabolism; Carcinoma/drug therapy; Carcinoma/metabolism; Carcinoma/pathology; Cell Adhesion/drug effects; Cell Adhesion/physiology; Cell Movement/drug effects; Cell Movement/physiology; Chemotaxis/physiology; Electrophoresis; Endothelium/physiology; Enzyme Activation; Humans; Ligands; Male; Matrix Metalloproteinase 2/metabolism; Matrix Metalloproteinase 9/metabolism; Mice; Mice, Nude; Molecular Sequence Data; Neoplasm Proteins/metabolism; Neoplasm Proteins/pharmacology; Neoplasm Proteins/physiology; Peptide Library; Prostatic Neoplasms/drug therapy; Prostatic Neoplasms/metabolism; Prostatic Neoplasms/pathology; Sequence Homology, Amino Acid; Substrate Specificity; Tumor Cells, Cultured |
References
|