-
Interaction
| AccNo. |
22516 |
Score |
0.86 |
| Name |
ARASE_hK2 |
Environment |
in vitro |
| Kd |
1.0 |
Organism |
N/A |
| Function |
peptide is a substrate of kallikrein 2 |
|
Peptide
| AccNo. |
22400 |
| Name |
ARASE |
| Organism |
N/A |
| Constraint |
none |
| Sequence |
ARASE
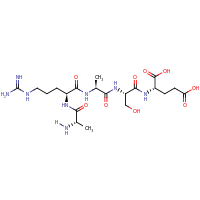
|
| Origin |
phage display |
| Form |
fused to cyan fluorescent protein, expressed in Escherichia coli |
| Internalized |
no |
| Unnatural |
no |
| Imaging |
no |
| Is Motif |
no |
|
Interactor
| AccNo. |
22250 |
| Name |
hK2 |
| Description |
kallikrein 2 |
| Organism |
Homo sapiens (human) |
| Type |
protein |
|
Experiment
| AccNo. |
22469 |
Classification incorrect?
Click the corresponding button to vote. Your vote
is used to improve the automatic classification.

|
| CA |
CVD |
DM |
APO |
ANG |
MI |
BD |
|
| 0.50 |
0.49 |
0.50 |
0.51 |
0.51 |
0.51 |
0.49 |
Vote |
 |
 |
 |
 |
 |
 |
 |
Yes |
 |
 |
 |
 |
 |
 |
 |
No |
|
| Name |
PD_6 |
| Detection |
protease assay, MI:0435 |
| Source |
PDF |
Text Id |
6 |
| Journal |
Eur J Biochem. 2002 Jun;269(11):2747-54. |
| Title |
Substrate specificity of human kallikrein 2 (hK2) as determined by phage display technology. |
| Authors |
Cloutier SM, Chagas JR, Mach JP, Gygi CM, Leisinger HJ, Deperthes D |
| Text |
Human glandular kallikrein 2 (hK2) is a trypsin-like serine protease expressed predominantly in the prostate epithelium. Recently, hK2 has proven to be a useful marker that can be used in combination with prostate specific antigen for screening and diagnosis of prostate cancer. The cleavage by hK2 of certain substrates in the proteolytic cascade suggest that the kallikrein may be involved in prostate cancer development; however, there has been very little other progress toward its biochemical characterization or elucidation of its true physiological role. In the present work, we adapt phage substrate technology to study the substrate specificity of hK2. A phage-displayed random pentapeptide library with exhaustive diversity was generated and then screened with purified hK2. Phages displaying peptides susceptible to hK2 cleavage were amplified in eight rounds of selection and genes encoding substrates were transferred from the phage to a fluorescent system using cyan fluorescent protein (derived from green fluorescent protein) that enables rapid determination of specificity constants. This study shows that hK2 has a strict preference for Arg in the P1 position, which is further enhanced by a Ser in P'1 position. The scissile bonds identified by phage display substrate selection correspond to those of the natural biological substrates of hK2, which include protein C inhibitor, semenogelins, and fibronectin. Moreover, three new putative hK2 protein substrates, shown elsewhere to be involved in the biology of the cancer, have been identified thus reinforcing the importance of hK2 in prostate cancer development. |
| Mesh Terms |
Green Fluorescent Proteins; Humans; Kinetics; Luminescent Proteins; Peptide Library; Substrate Specificity/genetics; Tissue Kallikreins/genetics; Tissue Kallikreins/metabolism |
References
|